The Databases of the USDA/ARS Plant Genome Research Program
Stephen R. Heller
USDA/ARS
Beltsville, MD 20705 USA
(srheller@gig.usda.gov)
and
Douglas W. Bigwood
Department of Plant Biology
University of Maryland
College Park, MD 20742 USA
(dbigwood@gig.usda.gov)
Abstract
This paper describes the creation and operation of a large domestic and international collaborative project in the area of plant genomics. Examples of how the system is made available via the World Wide Web are provided.
Introduction
Food is a basic need of mankind. With both a growing population and increasing
standards of living throughout the developed and developing countries of the world, the need for
increased supplies of food is apparent and will grow considerably in the decades ahead.
Biotechnology offers new tools for the agricultural scientist to help meet these needs.
Biotechnology can help address and solve problems of water supply, water quality, chemical
pesticide reduction, improved quality food products, biological controls of pests, and other
related issues.
In 1991 the United States Congress appropriated funds for the start of the USDA Plant
Genome Research Program, under the direction Dr. Jerome (Jerry) Miksche, to develop the
necessary tools to meet the food needs of the 21st century. The program consists of two basic
areas. The first area is for agricultural research efforts to map (and eventually, in some cases to
sequence) genes of agronomic importance, characterize these genes, and develop new plants with
improved traits of economic importance. The second area is to develop the necessary
infrastructure and support to assist the latter area. The system needed to support the handling
and manipulation of all this data is the heart of this project. Only by having quick, easy, and
accurate access to the vast amount of scientific data generated from the research results of
agricultural scientists around the world can one expect to need the food needs in the next century.
From the start of the project it was recognized that a large and sophisticated computer
system would be needed. Also, it was clear that the project needed to be global in scale, since
food is a universal need, and US agriculture contributes significantly to the US economy and
sales of US agriculture products are one of the few items for which there is a positive trade
surplus. Thus, the database and computer system effort was initiated at the onset, along with the
scientific research. Cooperation and collaboration on a large scale, with diverse groups of
scientists, virtually none of whom work for the same organization, is a difficult matter.
However, one of us (SRH) had experience developing a similar multi-organizational project, the
NIH/EPA Chemical Information System (1,2) and the experience from this project was used as
the basis for developing the computer system for the Plant Genome program.
The Plant Genome Database (PGD)
The first step in the database effort for this project was to find the appropriate
organization with the USDA to house the database. With an eye on the future of what libraries
will be in the next century, it was decided to locate the database activities at the US National
Agriculture Library (NAL), one of three (3) national libraries in the USA. This decision
agreement was easily made owing to the very positive attitude of the senior management of NAL
towards this project. The Department of Plant Biology at the University of Maryland was
awarded a contract to manage the project and develop the system.
Within months of the initiation of the project the authors were hired to begin the database and computer system development. One of us (SRH) was given responsibility for the overall management of the database, coordination of the researchers funded by USDA, and not funded by USDA (our collaborators). The other (DWB) was given the responsibility to manage a team of programmers to develop and make operational the actual computer systems. The project was both helped and hindered by the revolution in computer networking during the formative stages of the project. When the project first started the Internet was in its infancy and the World Wide Web (WWW) did not exist. As these technologies began to develop and emerge it made many of our efforts outdated very quickly. A more detailed history of the database development is available elsewhere (4). Details about the main software used in the Plant Genome project, ACEBD, is also available elsewhere (5,6) The focus of this paper will be on the current WWW implementation of the database.
AGIS
The system, which is now operational at the National Agriculture Library (NAL) and
available at no cost to the world-wide agriculture genome community, is called the Agricultural
Genome Information Server (AGIS). The home page of the system which has been developed
can be seen on the next page:
A Point Top 5% Site
and A Magellan 4 Star SiteWelcome to the Agricultural Genome Information Server sponsored by the U.S.
Department of Agriculture, Agricultural Research Service. AGIS is a cooperative effort
between the University of Maryland, College Park, Department of Plant Biology and the
National Agricultural Library. Browse and search the National Genetic Resources Program
genome databases and related biological information.
What's New? (30 October 1996) | Help | Search AGIS Docume nt Collection | How to... | Resources and Credits | Services ]
Figure 1. Plant Genome WWW AGIS Home Page
As seen in Figure 1, there are a number of items available in the Plant Genome Database
system. The first are the Databases, then there are Documents, listings of Conferences and
Meetings of interest to the project, Tools, and lastly, other methods of accessing and obtaining
data and information from the projects - namely by ftp and gopher. We will go through the
major items and show examples of each in order to present the reader with a basic understanding
of the size, scope, and amount of information available in the overall system.
In the WWW version of this paper (7) one can just click on the appropriate hypertext link
and go to the desired information. Thus, by clicking on the word "Genome" (found directly
under the heading : Databases) one goes to next page, shown in Figure 2, which shows all the
databases currently in the system.
[ What's New? (30 October 1996) | Help | Search AGIS Docum
ent Collection | How to... |
Resources and Credits | Services ]
Search Databases
[Plant Genome | Animal Genome | Other Genome | Plant Reference | Insect Reference]
Figure 2. Plant Genome Databases and Documents
By clicking on the words "browse", "query", or "about" one then is able to either browse
or search the database for particular information about the species chosen. By clicking on the
word "about" one is able to learn more about the database, such as the name of the curator, where
the database is being developed and maintained, and so on, before proceeding on to look at the
actual database. The browsing capability allows one to look at entries in the database for the
various classes or fields which are present. The number of entries for each class or field is also
given in the main listing.
For example, in the case of RiceGenes, there are 32 data classes from which one can chose.
They are shown in Figure 3 (October 1996 version of the system), one simply highlights the item
of interest, clicks on it, and the computer system goes directly that information.
Allele (579)
Author (2895)
Chrom_Block (137)
Colleague (260)
DNA (2154)
Definitions (5)
Gene_product (174)
Germplasm (377)
Homoeology (69)
Image (287)
Isolate (7)
Journal (73)
Keyword (2966)
Locus (3314)
LongText (23)
Map (100)
Map_Data (10)
Method (3)
Motif (339)
MultiMap (39)
Paper (1466)
Pathology (36)
Polymorphism (2376)
Probe (2403)
QTL (42)
Sequence (2780)
Species (34)
Text (1083)
Trait (1)
Var_release (23)
View (5)
gMap (14)
Figure 3. List of Classes or Fields of information in the Ricegenes database.
As an example, we have selected QTL as the data class and then retrieved a particular
data object: QTL qBlast-1-1. The result of this browsing is shown in Figure 4. The "view
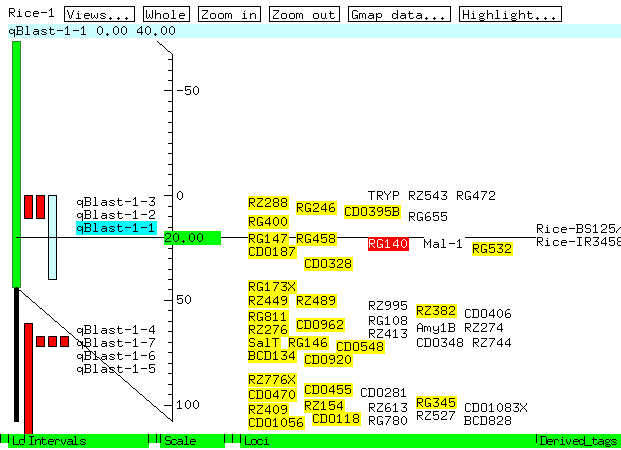
graphic" allows one to click here and go directly to a plot of the chromosome and the (labeled)
markers on the chromosome (as can be seen in Figure 5).
[view graphic]
Position Map Rice-1 Ends Left 0
Right 40
Positive Contains RG140
RG612
RED
Significant_loci RG612
Trait Blast-1
Location Greenhouse, Philippines
LOD_score 11.9
%_Variation_explained 32.5
Allele_effect 0.38
QTL_study QTL-Blast-CO39/Moroberekan
QTL_Remarks QTL for blast lesion number at LOD 6.0.
Markers RG140 and RG612 define the most
probable region. The most significant
marker was RG612, and statistics given
are for this marker only. Allele_effect
refers to the mean difference in lesion
number between the two genotypic groups
carrying the CO39 (susceptible) and
Moroberekan (resistant) alleles at the
significant_loci marker. Note that this
marker does not appear on the
Rice-BS125/2/BS125/WLO2 map, but is the
last marker on chromosome 1 on the map
produced by this QTL study.
Figure 4. Details of the RiceGenes QTL entry for qBlast-1-1.
Now, by clicking on "view graphic", one gets the actual map,
as shown below in Figure 5.

Figure 5. Graphic view of qBlast-1-1 QTL.
The graphics displays (which generate genetic maps, physical maps,
and sequences) are generated on-the-fly as GIF images.
They are scrollable, click-able, and zoom-able. If you click
on the name of a data object (as was shown in Figure 6), then the text of that data will be
retrieved. One drawback to the current display methods is that a new GIF must be generated for
each action. A new interface, written in the Java programming language, will provide the user
with a much more interactive environment.
To look at details of the trait information in Figure 4, one would click on the word "Blast-1" to the right of the word Trait and the following (Figure 6) appears:
Name Blast resistance
Evaluated_in QTL-Blast-CO39/Moroberekan
Description This study evaluated both qualitative
and quantitative resistance to rice
blast disease. For qualitative
resistance, the disease reaction in the
greenhouse was scored using a modified
scoring system based on the 0-5 scale.
An additional score of 3+ was added
between scores 3 and 4. Round lesions
of about 1-2 mm with gray centers and
brown margins and capable of sporulation
were classified as 3. Those with
round/elliptical lesions of about 2-3 mm
with gray centers and brown margins and
capable of sporulation were scores as
3+.
Three individual parameters of partial
resistance were measured during the
greenhouse phase of this study: diseased
leaf area, number of lesions, and size
of lesions. These 3 traits were
evaluated on 4 replications of 131 lines
using the blast isolate PO6-6, and were
used to identify QTL associated with
partial resistance to blast. These 3
components were significantly
correlated, and most of the markers
identified in the study affected all 3
parameters of blast. For field tests, a
randomized complete block design was
used. Two rows of a resistant cultivar
separated test entries, and 3 rows of
susceptible cultivars were planted
around the blocks. The percentage of
diseased leaf area was evaluated
visually at 3, 4, 5 and 6 weeks after
sowing. In Cavinti, two trials with two
replications were done. In Sitiung, one
trial with two replications was done.
The Allele_effect values given for the
QTLs refer to the mean differences in
either lesion number or percentage of
diseased leaf area between the two
genotypic groups carrying the CO39 and
the Moroberekan alleles at the
significant locus marker. This value
may be followed by links to specific
allele records. In this case, the first
allele is the one which increased the
trait of blast resistance, the second
allele decreased blast resistance.
Figure 6. Blast-1 trait details
While this short presentation does has not allowed for a full description of the many pieces of information in the different plant map databases, it is hoped this material has shown some of the content and capabilities of the system. With the database system being accessed over 200,000 times per month, it is clear that it is a very useful system. The full scope and impact of the system can only be assess by visiting and exploring the system (8).
Acknowledgment
The authors would like to thank Dr. Jerry Miksche for his help and encouragement in
developing the database and his leadership in the project. Without him this database would still
be only a plan, not a true living and used system.
References
(1) S. R. Heller, G. W. A. Milne, and R. J. Feldmann, "A Computer Based Chemical
Information System", Science, 195, 253-259(1977).
(2) G. W. A. Milne, R. Potenzone Jr., and S. R. Heller, "Environmental Uses of the NIH-EPA
Chemical Information System", Science, 215, 371-375(1982).
(3) The other two are the Library of Congress and the National Library of Medicine.
(4) "USDA Plant Genome Research Program, Advances in Agronomy, Volume 55, 113-166
(1995). In particular, see pages 147-154.
(5) J. M. Cherry, and S. W. Cartinhour, ACEDB, a tool for biological information. In Automated
DNA Sequencing and Analysis, Edited by M. Adams, C. Fields, and C. Venter, 347-356 (1994).
Academic Press: San Diego, CA.
(6) R. Durbin and J. T. Mieg (1991-). A C. elegansDatabase. Documentation, code and data
available from anonymous FTP servers at lirmm.lirmm.fr, cele.mrc-lmb.cam.ac.uk and
ncbi.nlm.nih.gov.
(7) A WWW version of this paper is available by pointing your browser to the url:
http://probe.nalusda.gov:8000/codata.html
(8) The AGIS system is available by pointing your browser to the url:http://probe.nalusda.gov:8000